
January 18, 2023
When I first began using ChatGPT, the latest implementation of GPT 3 from OpenAI, the phrase “gifted child” is what first came to mind. A problematic phrase, I know. While the anthropomorphization of artificial intelligence tends to cause us real humans to systematically over- or underestimate an algorithm’s abilities, that seems to be exactly what the folks over at OpenAI want us to do: chat with it. Like a child who is desperate to live up to the ever unfortunate “gifted” moniker, it seems almost desperate–yet strangely confident–to tell me what I want to hear, even when it’s factually wrong. However, very unlike any human being I’ve ever met, it also seems to have vast knowledge of just about any topic under the sun.
With all of that predictive power under the hood, I wanted to know whether this technology could be used to augment the systematic review process. By asking it a series of questions and prompting it to undertake a series of tasks related to systematic reviews and comprehension of clinical literature, I was able to provoke some very interesting responses.
I started where any literature review starts–by trying to construct a Search. I wanted to see if ChatGPT could generate a PubMed query that would yield similar initial search results to a human’s initial search string. I figured it would be easiest to simply try to replicate the initial search from a recently published review conducted by our research team in collaboration with two physicians from Stanford. The initial query looks like this:
basilar AND (stent-triever OR aspiration OR thrombectomy) AND (IVT OR IV-tPA OR thrombolysis) AND stroke
This query returns 260 results at the time of this writing, a very manageable result to screen. In an attempt to get ChatGPT to produce a query that returns a similar result, I gave it the following prompt:

Create a pubmed query that returns literature on basilar artery strokes and compares thrombectomy to thrombolysis.
To which it responded:

Here is an example of a Pubmed query that could be used to search for literature on basilar artery strokes and compare thrombectomy to thrombolysis:
("basilar artery stroke"[MeSH Terms] OR "basilar artery stroke"[All Fields]) AND ("thrombectomy"[MeSH Terms] OR "thrombectomy"[All Fields] OR "thrombolysis"[MeSH Terms] OR "thrombolysis"[All Fields]) AND ("comparative study"[Publication Type] OR "randomized controlled trial"[Publication Type])
This query searches for articles that have "basilar artery stroke" in the MeSH terms or in any field, and have "thrombectomy" or "thrombolysis" in the MeSH terms or any field, and are either a "comparative study" or a "randomized controlled trial" in the publication type.
Please note that this search query not guaranteed to return a perfect results, as you may have to modify the query depending on your specific research needs.
Not bad, but not great. Here are some simple pros and cons regarding ChatGPT’s performance:
Pros:
Cons:
So, while the syntax and basic concepts are there, the problems of ChatGPT making up MeSH terms is major. Scientific search depends on replicable, consistent query construction, and MeSH is a controlled vocabulary, not just a modifier to append to any concept.
Thankfully, it has also included “basilar artery stroke”[All Fields] as well, so at least that term should still yield some results. However, because Chat GPT is rather arbitrarily throwing quotes around “basilar artery stroke” making PubMed look for exact matches only, this search returned only two studies. While Nested Knowledge’s systematic review does eventually winnow our 260 results down to four RCTs of interest, this search only contains one of those studies.
I tried several times to address these issues by asking ChatGPT similar questions to try to get a larger set of results. After several attempts that did nothing to address the basic issue, I threw up my hands and asked: “Can you broaden these results further? This query only returns two results.”
And it complies:

I apologize, it's possible that the query I provided is too specific, which resulted in a small number of results. Here are a few suggestions that may help broaden your search results:
Remove the MeSH terms from your query:
MeSH terms are assigned to articles by indexers at the National Library of Medicine and may not always be present in all relevant articles. Removing the MeSH terms from the query will allow you to search for the terms in any field.
Use a broader term in place of "basilar artery stroke":
Instead of searching for "basilar artery stroke" specifically, you could use a broader term such as "cerebrovascular accident" or "stroke" to capture more articles.
Remove the specific thrombectomy techniques from the query:
Instead of searching for specific thrombectomy techniques like "stent-triever" or "aspiration", you could search for thrombectomy in general, so you can get a more general overview of the thrombectomy and it's indications.
Increase the date range of your search:
If you increase the date range of your search, you may be able to find more recent articles that were not included in your original search.
Change the order of the words of the query:
Try altering the order of the words of the query can change the results you're getting, for example by rearranging the terms.
Search for related terms:
Look for related terms and synonyms that might be used in the literature, and include them in your query.
Here is an example of a modified query that incorporates these suggestions:
(stroke OR cerebrovascular accident) AND thrombectomy AND "randomized controlled trial"[Publication Type]
This query will look for articles that match either of the terms "stroke" or "cerebrovascular accident" and "thrombectomy", and are "randomized controlled trial" in the publication type. It's possible that this query will bring up a larger number of results, but please keep in mind that it will not be as specific as the previous query.
Still not perfect, (thrombolysis is notably absent now) but this query does actually return the four RCTs that I know should be included in this review, somewhere amongst 295 records, which is similar in size to my example query. I’m not sure this process was faster or easier than just writing that query by hand, especially for a more seasoned research librarian than myself. I did find some of the ideas it gave me to be useful, such as including “randomized controlled trial” as a publication type, or to feeding me synonyms, like cerebrovascular accident. However, my experiment here is obviously contrived; any search query created by ChatGPT is only going to be as accurate as the doctor who ordered it, and will require nearly as much input and brainpower as simply writing the query the old fashioned way.
After watching ChatGPT make up MeSH terms, I am apprehensive about letting it handle tasks alone. However, identifying relationships between words is actually something that Large Language Models (LLMs) specialize in, which is similar to tagging in Nested Knowledge. While we do have a “Tag Recommendations” feature, it currently only looks for matches tag names or aliases of tags in full texts. With ChatGPT’s ability to predict relationships and identify context, it might be possible to make our Tag Recommendations feature much smarter, even if the accuracy is not perfect.
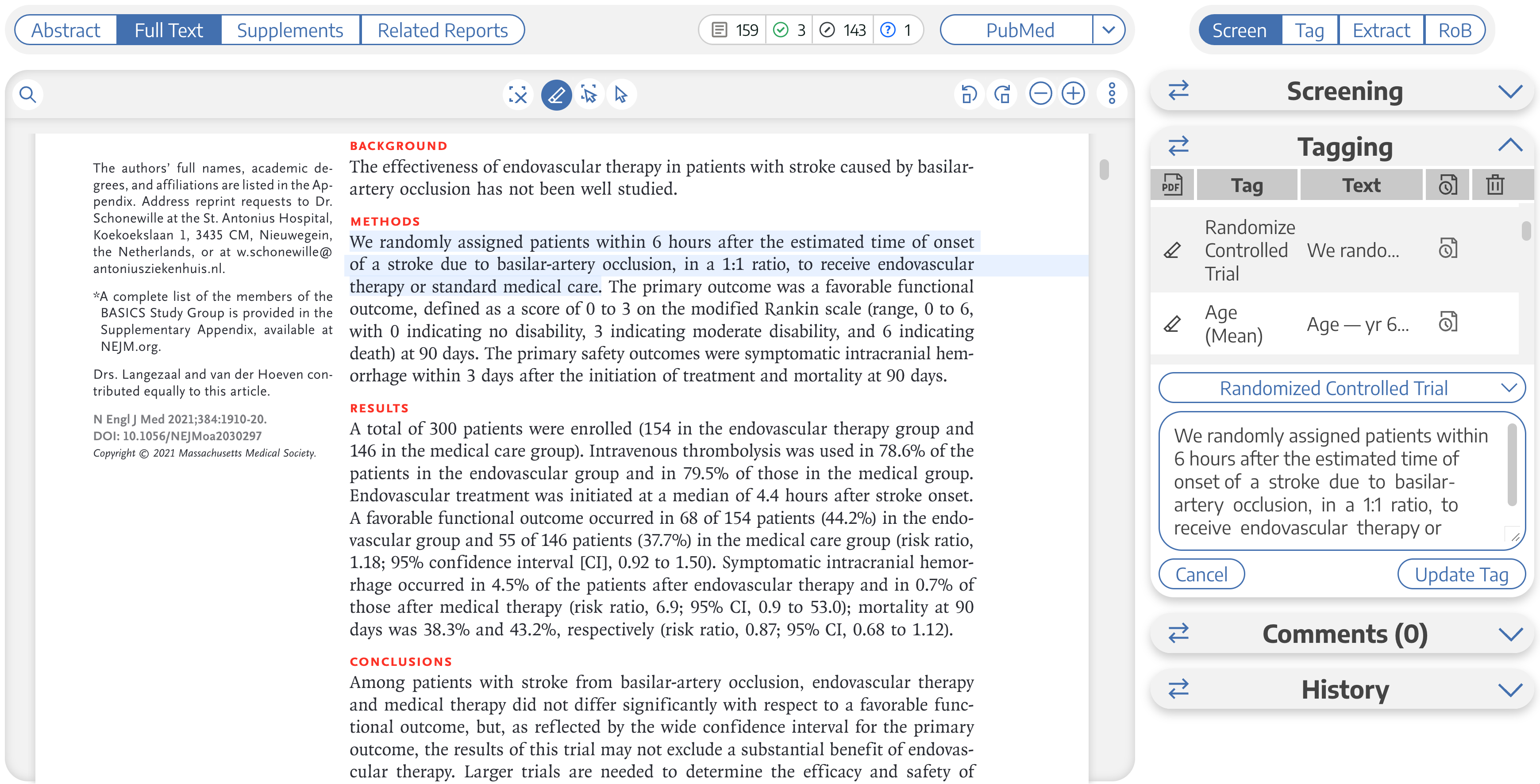
To test its abilities, I began by attempting to copy and paste the entire full text of one of the four aforementioned RCTs into ChatGPT…only to immediately be met by an error. Both the prompt and the conversation history together are limited to about 3000 words (4000 tokens). However, it did accept the abstract, which we know contains the “Randomized Controlled Trial” tag.
In an effort to simulate tagging this abstract in ChatGPT, I prompted:

Create a pubmed query that returns literature on basilar artery strokes and compares thrombectomy to thrombolysis.
It managed to identify the same portion of the text as our human reviewer:

We randomly assigned patients within 6 hours after the estimated time of onset of a stroke due to basilar-artery occlusion, in a 1:1 ratio, to receive endovascular therapy or standard medical care.
An impressive result, to be sure, but admittedly still contrived, given that I had to feed it less than the full text. It’s also a bit of a softball, because getting to the right answer doesn’t require parsing as much context as some of the other tags in this nest. So let’s play hardball.
In this particular study, some of the data we need to tag was formatted as a table, which while not uncommon, requires apprehending a lot more context in order to find the right answer. Merely copying and pasting this table into ChatGPT strips out a lot of the formatting that makes tabular data so readable for human beings.
But here too, ChatGPT doesn’t even flinch:

The table you provided presents the baseline characteristics of the patients in the study. It lists various demographic and clinical characteristics of the patients assigned to the endovascular therapy group and the medical care group. It includes information such as the patients' age, gender, medical history, prestroke functional status, treatment with intravenous thrombolysis, time from stroke onset to treatment, and stroke severity. The table also includes information on the reperfusion status on digital subtraction angiography. The table provides data on 154 patients in the endovascular therapy group and 146 patients in the medical care group.

Not only did it correctly identify the structure of the content, it seemed to have no issue lining up the table’s cells with the appropriate labels in both columns and rows. As a way to get it to “tag” for age I asked:

Find Age (Mean)

The mean age of the patients in the endovascular therapy group is 66.8 years, and in the medical care group is 67.2 years.
Again, it’s taking my very short query a little too literally by giving me only the mean and leaving out the standard deviation, but that’s easily remedied:

Can you also give me the standard deviation for those averages?

The mean age of the patients in the endovascular therapy group is 66.8 years (SD 13.1) and in the medical care group is 67.2 years (SD 11.9).
Whether or not this could perform at this level of accuracy for every data element across all the studies in a typical meta-analysis remains to be seen, but this initial result seems to indicate that this technology could be used to dramatically speed up quantitative extraction, and even begin to do some of the work for the researcher.
For now at least, GPT 3 is in its infancy, but already I see the eerie promise of an almost gifted, almost child. Like a child, it has soaked up 175 billion parameters in the first few months of life. Now, it’s parsing, matching, sorting each bit of information in order to predict and mimic the correct responses to all of my questions, as I attempt to teach it how to operate and adapt to my very particular context. With GPT 4 set to launch some time in late 2023, I am reminded that children tend to grow up, often faster than we expect.
As this technology matures, its predictions and mimicry could begin to align so closely with the truth that we just start calling them accurate. Certainly, AI is already surpassing the accuracy of humans at certain tasks. However, I feel it’s important to remember that this tool is still just an algorithm, meaning it will always require human input. In the realm of systematic reviews especially, human expertise is needed to provide the initial search concepts of interest, identify qualitative concepts we want to contrast across studies, and indicate the quantitative data elements for which we want to compute treatment effects. The tools at hand may evolve, but it’s always going to be the human researcher who dreams up and structures the study.
With the promising results in mind, we at Nested Knowledge will certainly consider ways to incorporate AI like this into our software in places where perfect accuracy matters less, or where the results of automation can be checked for accuracy by reviewers. It’s very possible that ChatGPT or something like it could integrate directly into our platform and offer more intelligent suggestions, better predictions, and quick summaries in the very near future.